Our projects on deep Learning for Biomedicine
This front adapts from our legacy website
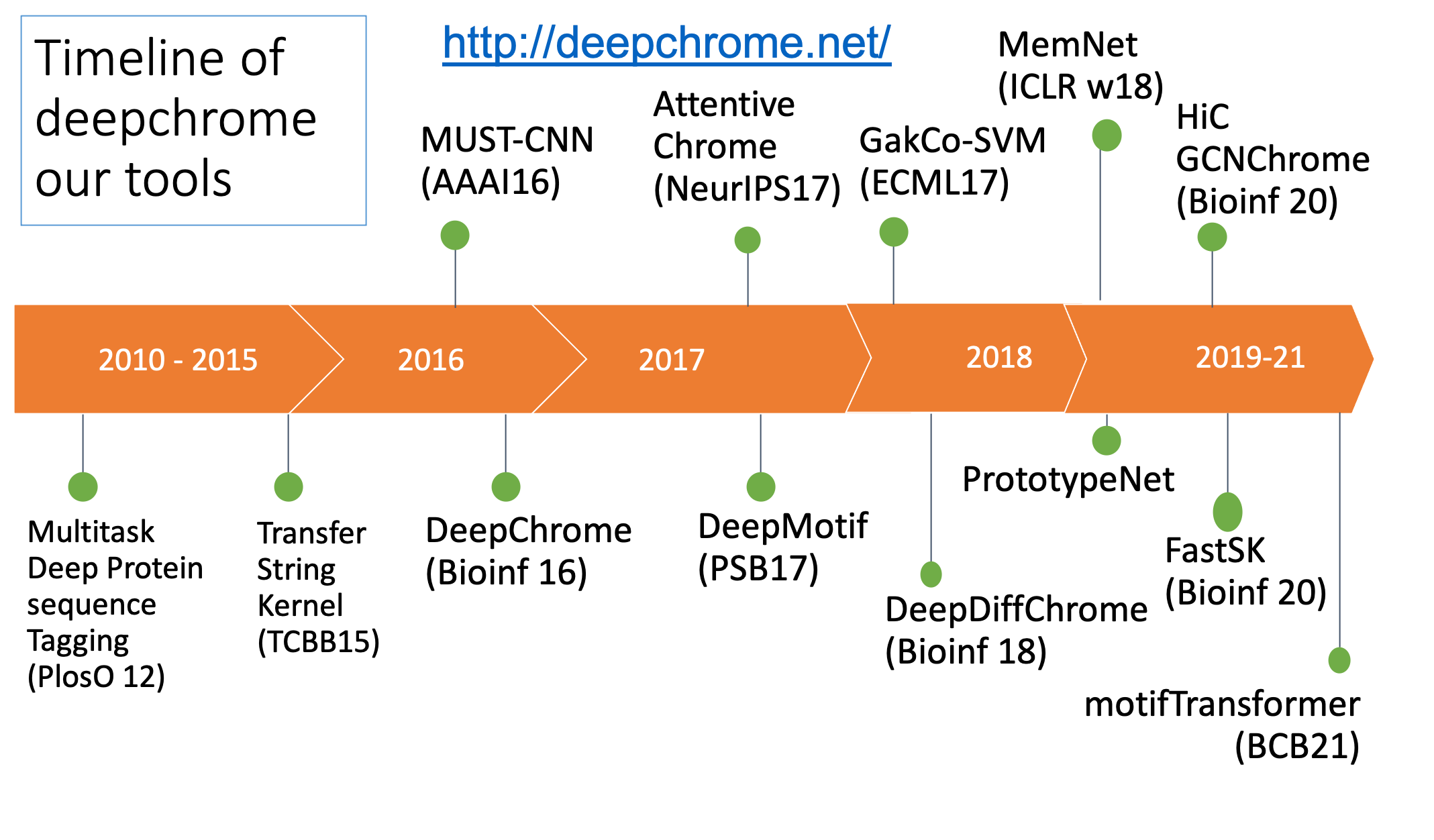
deepchrome.org (later to deepchrome.net) and introduces updates of a suite of
deep learning tools we have developed for learning patterns and making predictions on biomedical data (mostly from functional genomics).
Please feel free to email me when you find my typos.
General Background
Biology and medicine are rapidly becoming data-intensive. A recent comparison of genomics with social media, online videos, and other data-intensive disciplines suggests that genomics alone will equal or surpass other fields in data generation and analysis within the next decade. The volume and complexity of these data present new opportunities, but also pose new challenges. Data sets are complex, often ill-understood and grow at a a faster scale than computational capabilities. Problems of this nature may be particularly well-suited to deep learning techniques (see Opportunities and obstacles for deep learning in biology and medicine). The website introduces a suite of deep learning tools we have developed for learning patterns and making predictions on biomedical data (mostly from functional genomics).
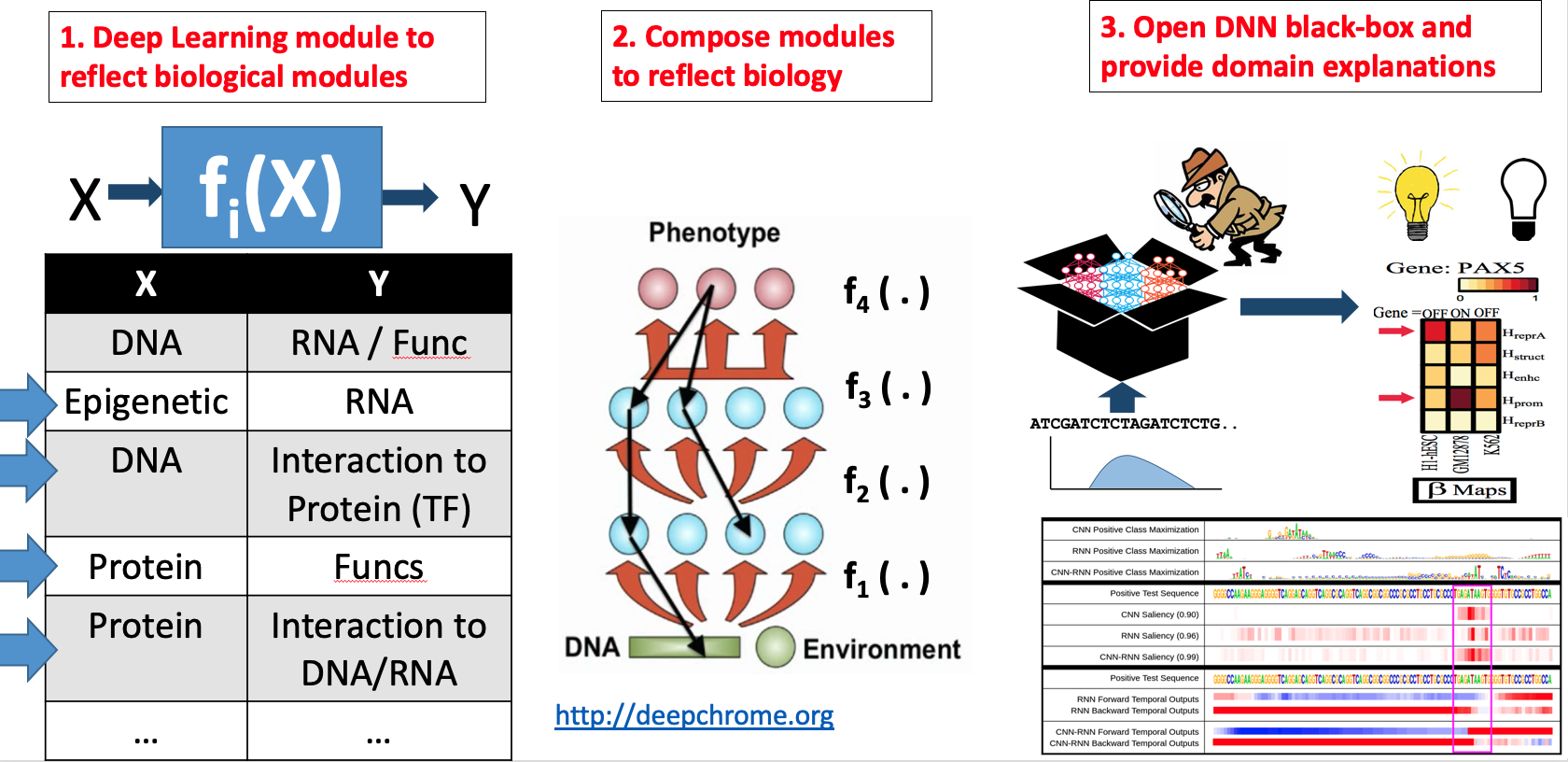
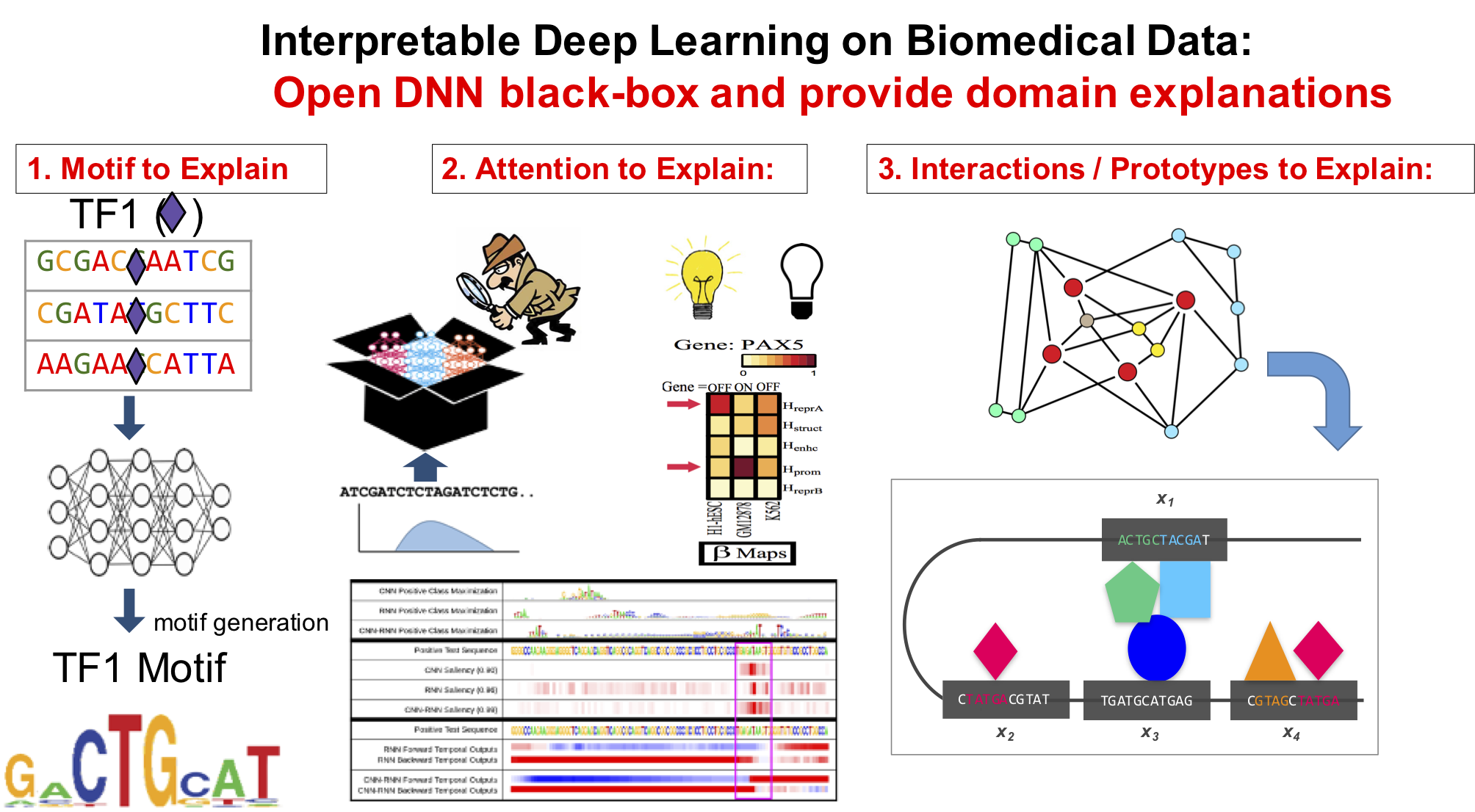
Our technical focus in this direction center on making DNN interpretable.
Background of Learning: Representation Learning and Deep Learning
The performance of machine learning algorithms is largely dependent on the
data representation (or features) on which they are
applied. Deep learning aims at discovering learning algorithms that can
find multiple levels of representations directly from data, with higher
levels representing more abstract concepts. In recent years,
the field of deep learning has lead to groundbreaking performance in many applications such as computer vision, speech understanding, natural language processing, and computational biology.
Have questions or suggestions? Feel free to ask me on Twitter or email me.
Thanks for reading!
less than 1 minute read
I gave a tutorial talk at UVA-VADC Seminar Series 2021 and at monthly NIH Data Science Showcase seminar.
less than 1 minute read
I gave a tutorial talk at
UVA-CPHG Seminar Series 2018.
1 minute read
Tool DeepDIff: DeepDiff: Deep-learning for predicting Differential gene expression from histone modifications
less than 1 minute read
Here are the slides of tutorial talk I gave at ACM-BCB 2018.
1 minute read
Ph.D. Dissertation Defense by Ritambhara Singh
Monday, April 9, 2018 at 12:00PM in Rice 504.
less than 1 minute read
Here are the slides of lecture talks I gave at UCLA CGWI and NLM-CBB seminar about our deep learning tools: DeepChrome, AttentiveChrome and DeepMotif.
less than 1 minute read
Jack’s DeepMotif paper (Deep Motif Dashboard: Visualizing and Understanding Genomic Sequences Using Deep Neural Networks ) have received the “best paper awar...
1 minute read
Tool AttentiveChrome: Attend and Predict: Using Deep Attention Model to Understand Gene Regulation by Selective Attention on Chromatin
1 minute read
Tool Deep Motif Dashboard: Visualizing and Understanding Genomic Sequences Using Deep Neural Networks
1 minute read
Tool DeepChrome: deep-learning for predicting gene expression from histone modifications
1 minute read
Tool GaKCo-SVM: a Fast GApped k-mer string Kernel using COunting
1 minute read
Tool TSK: Transfer String Kernel for Cross-Context DNA-Protein Binding Prediction
less than 1 minute read
Tool MUST-CNN: A Multilayer Shift-and-Stitch Deep Convolutional Architecture for Sequence-based Protein Structure Prediction
1 minute read
Title: Deep Learning for Character-based Information Extraction on Chinese and Protein Sequence
less than 1 minute read
Here are the slides of one lecture talk I gave at UVA CPHG Seminar Series in 2014 about our deep learning tools back then.
1 minute read
Tool Multitask-ProteinTagging: A unified multitask architecture for predicting local protein properties



