ACM BCB - Transfer Learning for Predicting Virus-Host Protein Interactions for Novel Virus Sequences
Title: Transfer Learning for Predicting Virus-Host Protein Interactions for Novel Virus Sequences
- authors: Jack Lanchantin, Tom Weingarten, Arshdeep Sekhon, Clint Miller, Yanjun Qi
- 2021 ACM Conference on Bioinformatics, Computational Biology, and Health Informatics (ACM BCB)
PDF @ BioArxiv
Talk: Slide
Abstract
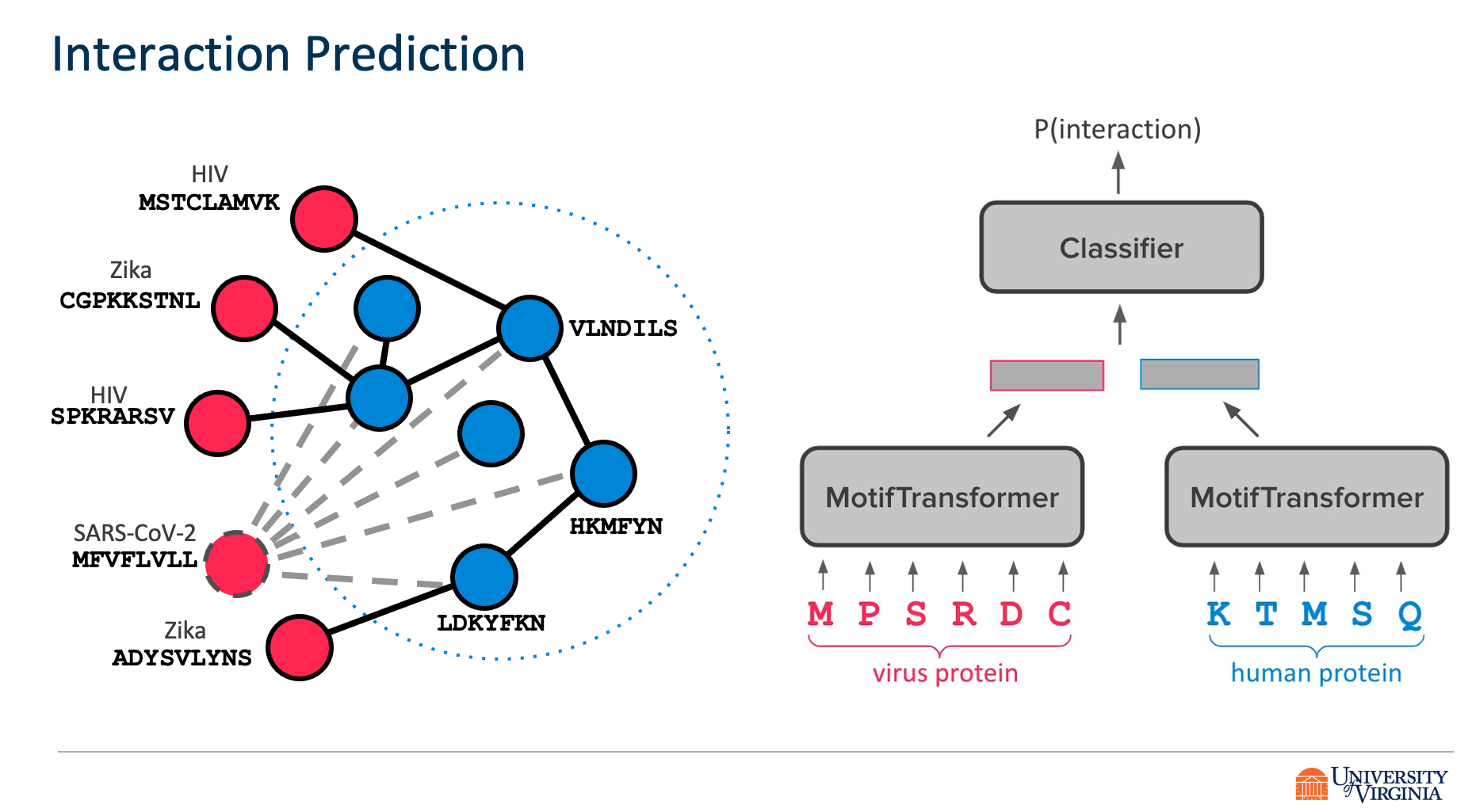
Viruses such as SARS-CoV-2 infect the human body by forming interactions between virus proteins and human proteins. However, experimental methods to find protein interactions are inadequate: large scale experiments are noisy, and small scale experiments are slow and expensive. Inspired by the recent successes of deep neural networks, we hypothesize that deep learning methods are well-positioned to aid and augment biological experiments, hoping to help identify more accurate virus-host protein interaction maps. Moreover, computational methods can quickly adapt to predict how virus mutations change protein interactions with the host proteins.
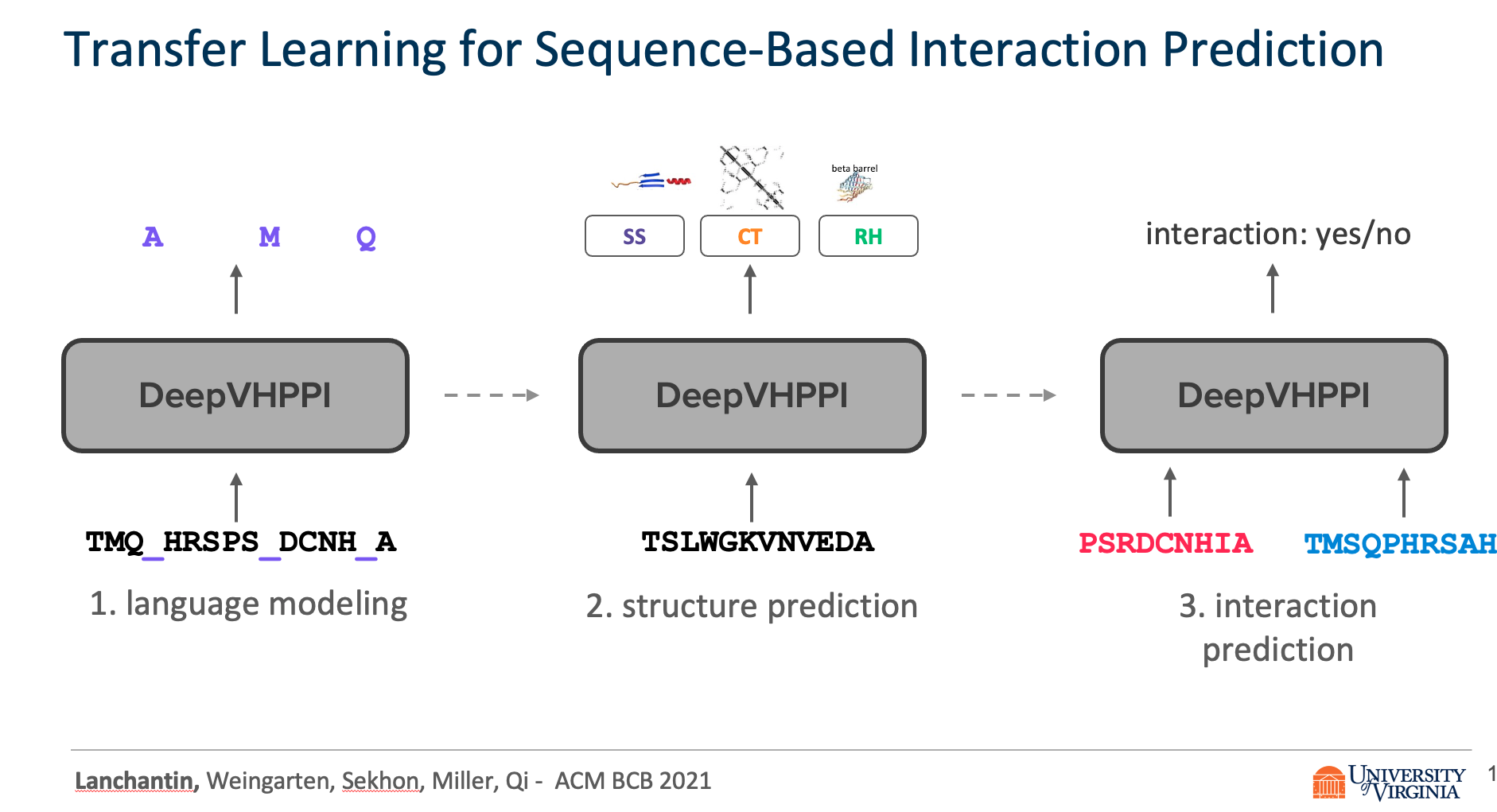
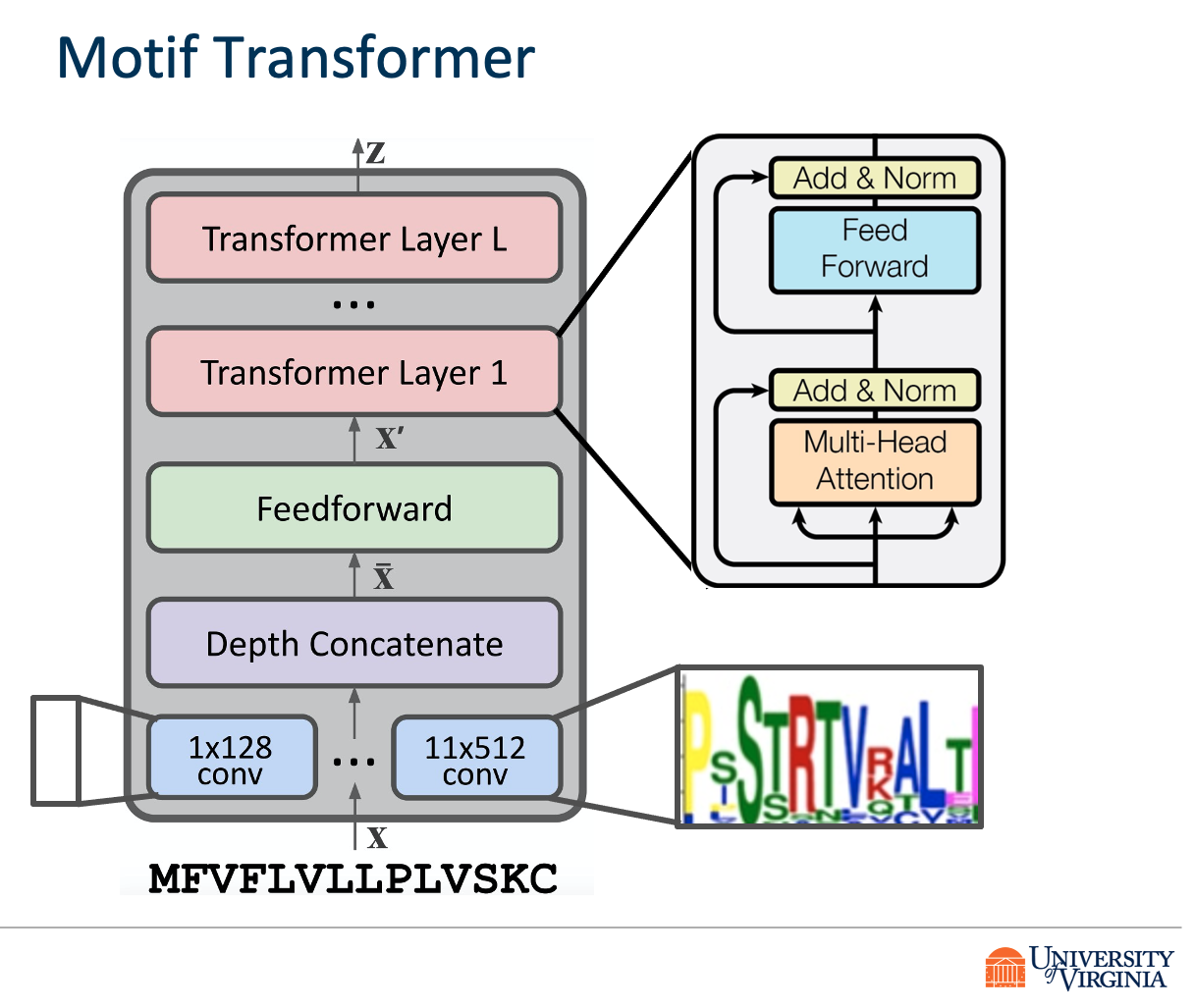
We propose DeepVHPPI, a novel deep learning framework combining a self-attention-based transformer architecture and a transfer learning training strategy to predict interactions between human proteins and virus proteins that have novel sequence patterns. We show that our approach outperforms the state-of-the-art methods significantly in predicting Virus–Human protein interactions for SARS-CoV-2, H1N1, and Ebola. In addition, we demonstrate how our framework can be used to predict and interpret the interactions of mutated SARS-CoV-2 Spike protein sequences.
We make all of our data and code available on GitHub https://github.com/QData/DeepVHPPI.
Citations
@article {Lanchantin2020.12.14.422772,
author = {Lanchantin, Jack and Weingarten, Tom and Sekhon, Arshdeep and Miller, Clint and Qi, Yanjun},
title = {Transfer Learning for Predicting Virus-Host Protein Interactions for Novel Virus Sequences},
elocation-id = {2020.12.14.422772},
year = {2021},
doi = {10.1101/2020.12.14.422772},
publisher = {Cold Spring Harbor Laboratory},
abstract = {Viruses such as SARS-CoV-2 infect the human body by forming interactions between virus proteins and human proteins. However, experimental methods to find protein interactions are inadequate: large scale experiments are noisy, and small scale experiments are slow and expensive. Inspired by the recent successes of deep neural networks, we hypothesize that deep learning methods are well-positioned to aid and augment biological experiments, hoping to help identify more accurate virus-host protein interaction maps. Moreover, computational methods can quickly adapt to predict how virus mutations change protein interactions with the host proteins.We propose DeepVHPPI, a novel deep learning framework combining a self-attention-based transformer architecture and a transfer learning training strategy to predict interactions between human proteins and virus proteins that have novel sequence patterns. We show that our approach outperforms the state-of-the-art methods significantly in predicting Virus{\textendash}Human protein interactions for SARS-CoV-2, H1N1, and Ebola. In addition, we demonstrate how our framework can be used to predict and interpret the interactions of mutated SARS-CoV-2 Spike protein sequences.Availability We make all of our data and code available on GitHub https://github.com/QData/DeepVHPPI.ACM Reference Format Jack Lanchantin, Tom Weingarten, Arshdeep Sekhon, Clint Miller, and Yanjun Qi. 2021. Transfer Learning for Predicting Virus-Host Protein Interactions for Novel Virus Sequences. In Proceedings of ACM Conference (ACM-BCB). ACM, New York, NY, USA, 10 pages. https://doi.org/??Competing Interest StatementThe authors have declared no competing interest.},
URL = {https://www.biorxiv.org/content/early/2021/06/08/2020.12.14.422772},
eprint = {https://www.biorxiv.org/content/early/2021/06/08/2020.12.14.422772.full.pdf},
journal = {bioRxiv}
}
Support or Contact
Having trouble with our tools? Please contact Jack and we’ll help you sort it out.
