18 Jun 2019
Abstract
We focus on integrating different types of extra knowledge (other than the observed samples) for estimating the sparse structure change between two p-dimensional Gaussian Graphical Models (i.e. differential GGMs). Previous differential GGM estimators either fail to include additional knowledge or cannot scale up to a high-dimensional (large p) situation. This paper proposes a novel method KDiffNet that incorporates Additional Knowledge in identifying Differential Networks via an Elementary Estimator. We design a novel hybrid norm as a superposition of two structured norms guided by the extra edge information and the additional node group knowledge. KDiffNet is solved through a fast parallel proximal algorithm, enabling it to work in large-scale settings. KDiffNet can incorporate various combinations of existing knowledge without re-designing the optimization. Through rigorous statistical analysis we show that, while considering more evidence, KDiffNet achieves the same convergence rate as the state-of-the-art. Empirically on multiple synthetic datasets and one real-world fMRI brain data, KDiffNet significantly outperforms the cutting edge baselines concerning the prediction performance, while achieving the same level of time cost or less.
Citations
@conference{arsh19kdiffNet,
Author = {Sekhon, Arshdeep and Wang, Beilun and Qi, Yanjun},
Title = {Adding Extra Knowledge in Scalable Learning of
Sparse Differential Gaussian Graphical Models},
Year = {2019}}
}
Having trouble with our tools? Please contact Arsh and we’ll help you sort it out.
12 May 2018
Paper: Most updated version at HERE | Previous version: @Arxiv |
TalkSlide: URL
R package: URL
GitRepo for R package: URL
install.packages("jeek")
library(jeek)
demo(jeek)
Abstract
We consider the problem of including additional knowledge in estimating sparse Gaussian graphical models (sGGMs) from aggregated samples, arising often in bioinformatics and neuroimaging applications. Previous joint sGGM estimators either fail to use existing knowledge or cannot scale-up to many tasks (large $K$) under a high-dimensional (large $p$) situation. In this paper, we propose a novel \underline{J}oint \underline{E}lementary \underline{E}stimator incorporating additional \underline{K}nowledge (JEEK) to infer multiple related sparse Gaussian Graphical models from large-scale heterogeneous data. Using domain knowledge as weights, we design a novel hybrid norm as the minimization objective to enforce the superposition of two weighted sparsity constraints, one on the shared interactions and the other on the task-specific structural patterns. This enables JEEK to elegantly consider various forms of existing knowledge based on the domain at hand and avoid the need to design knowledge-specific optimization. JEEK is solved through a fast and entry-wise parallelizable solution that largely improves the computational efficiency of the state-of-the-art $O(p^5K^4)$ to $O(p^2K^4)$. We conduct a rigorous statistical analysis showing that JEEK achieves the same convergence rate $O(\log(Kp)/n_{tot})$ as the state-of-the-art estimators that are much harder to compute.
Empirically, on multiple synthetic datasets and two real-world data, JEEK outperforms the speed of the state-of-arts significantly while achieving the same level of prediction accuracy.
About Adding Additional Knowledge
One significant caveat of state-of-the-art joint sGGM estimators is the fact that little attention has been paid to incorporating existing knowledge of the nodes or knowledge of the relationships among nodes in the models.
In addition to the samples themselves, additional information is widely available in real-world applications. In fact, incorporating the knowledge is of great scientific interest. A prime example is when estimating the functional brain connectivity networks among brain regions based on fMRI samples, the spatial position of the regions are readily available. Neuroscientists have gathered considerable knowledge regarding the spatial and anatomical evidence underlying brain connectivity (e.g., short edges and certain anatomical regions are more likely to be connected \cite{watts1998collective}). Another important example is the problem of identifying gene-gene interactions from patients’ gene expression profiles across multiple cancer types. Learning the statistical dependencies among genes from such heterogeneous datasets can help to understand how such dependencies vary from normal to abnormal and help to discover contributing markers that influence or cause the diseases. Besides the patient samples, state-of-the-art bio-databases like HPRD \cite{prasad2009human} have collected a significant amount of information about direct physical interactions among corresponding proteins, regulatory gene pairs or signaling relationships collected from high-qualify bio-experiments.
Although being strong evidence of structural patterns we aim to discover, this type of information has rarely been considered in the joint sGGM formulation of such samples. This paper aims to fill this gap by adding additional knowledge most effectively into scalable and fast joint sGGM estimations.
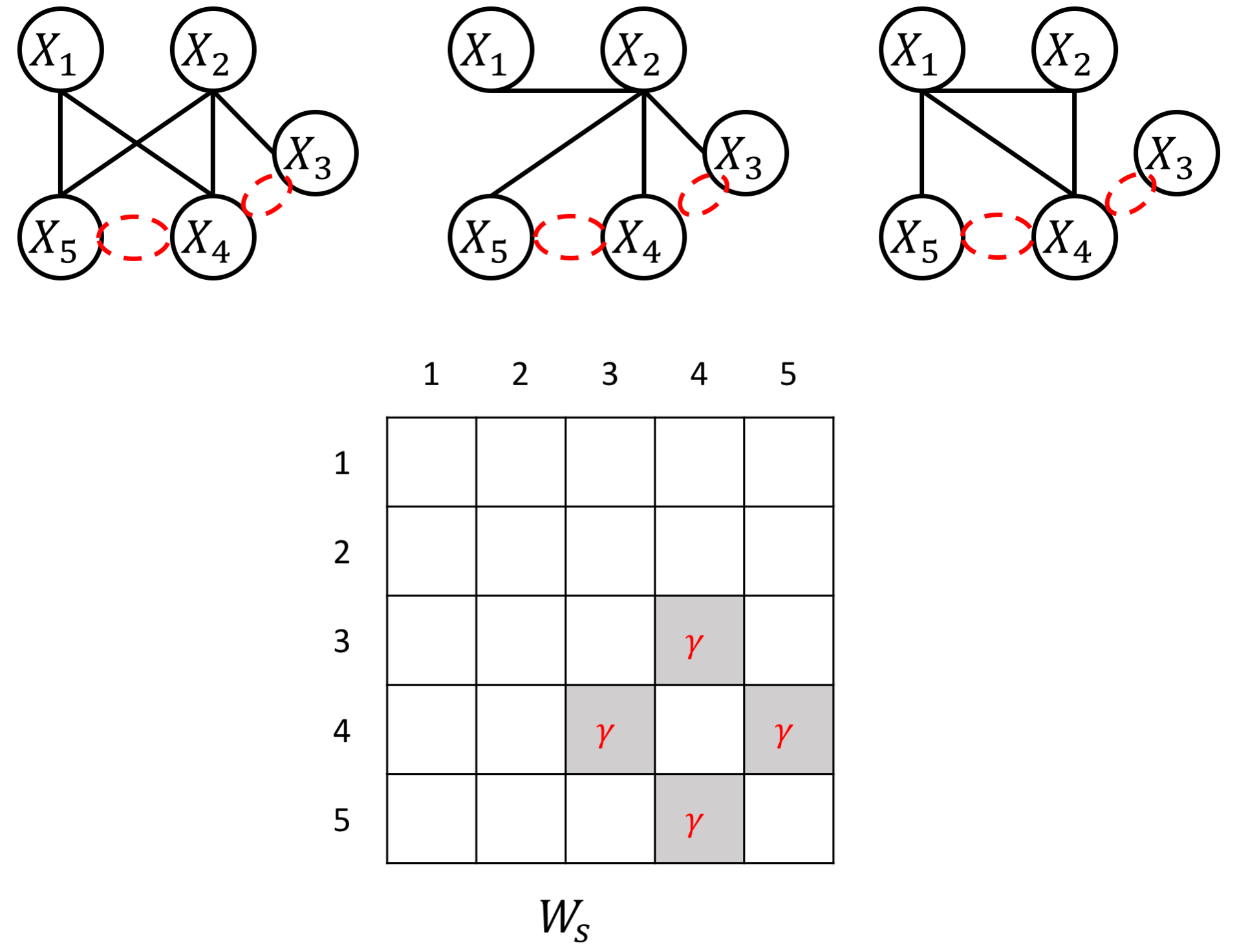
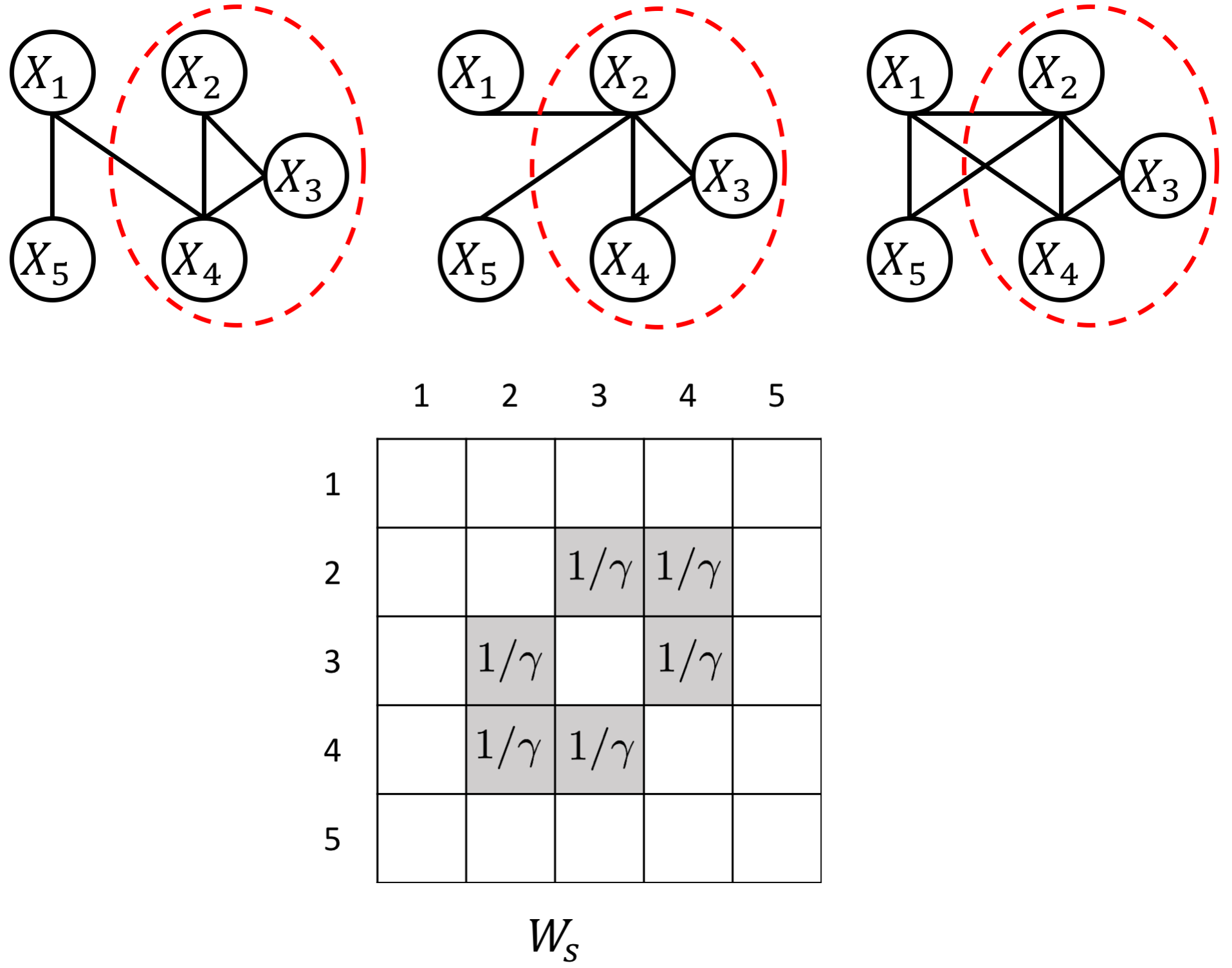
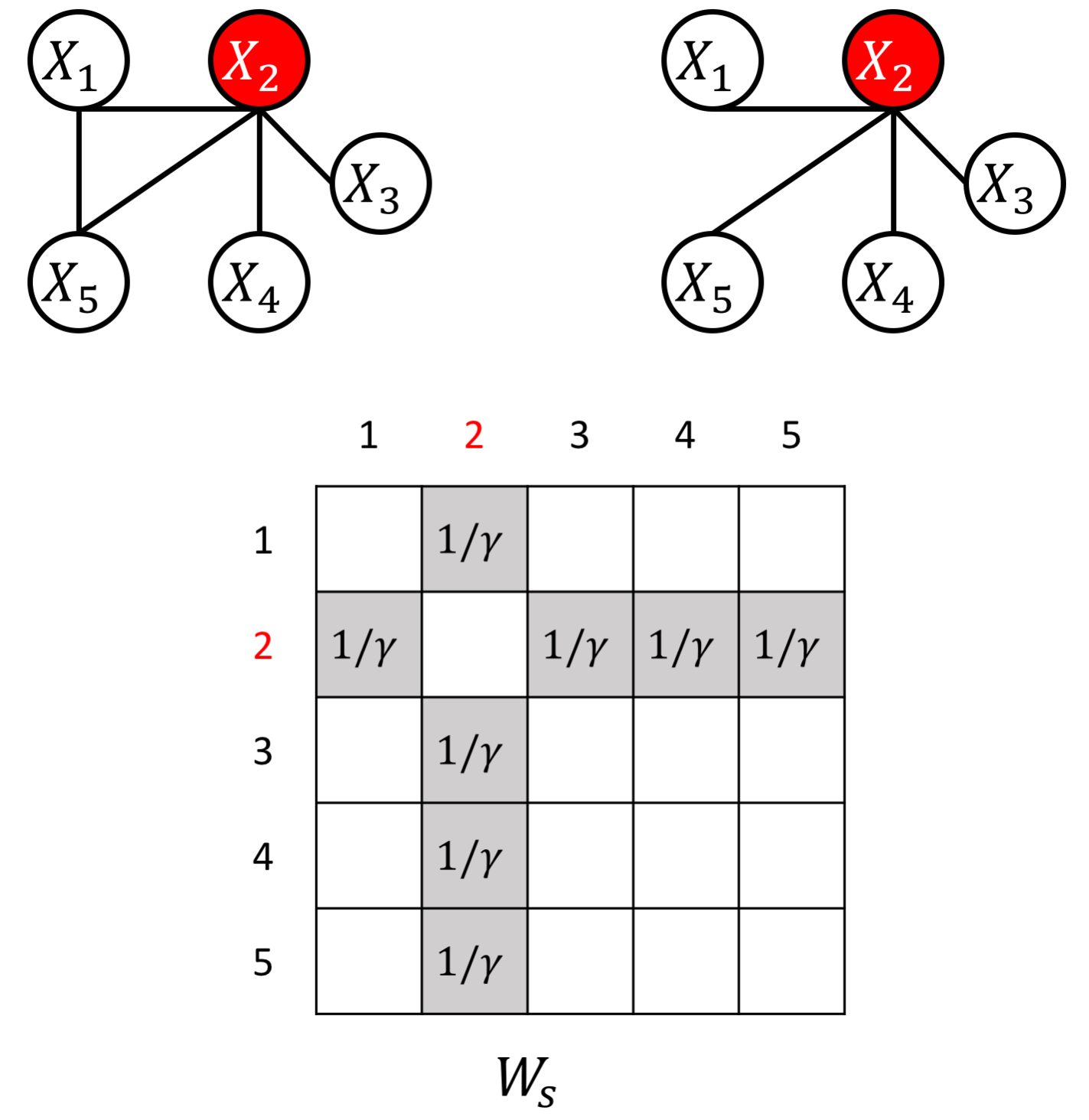
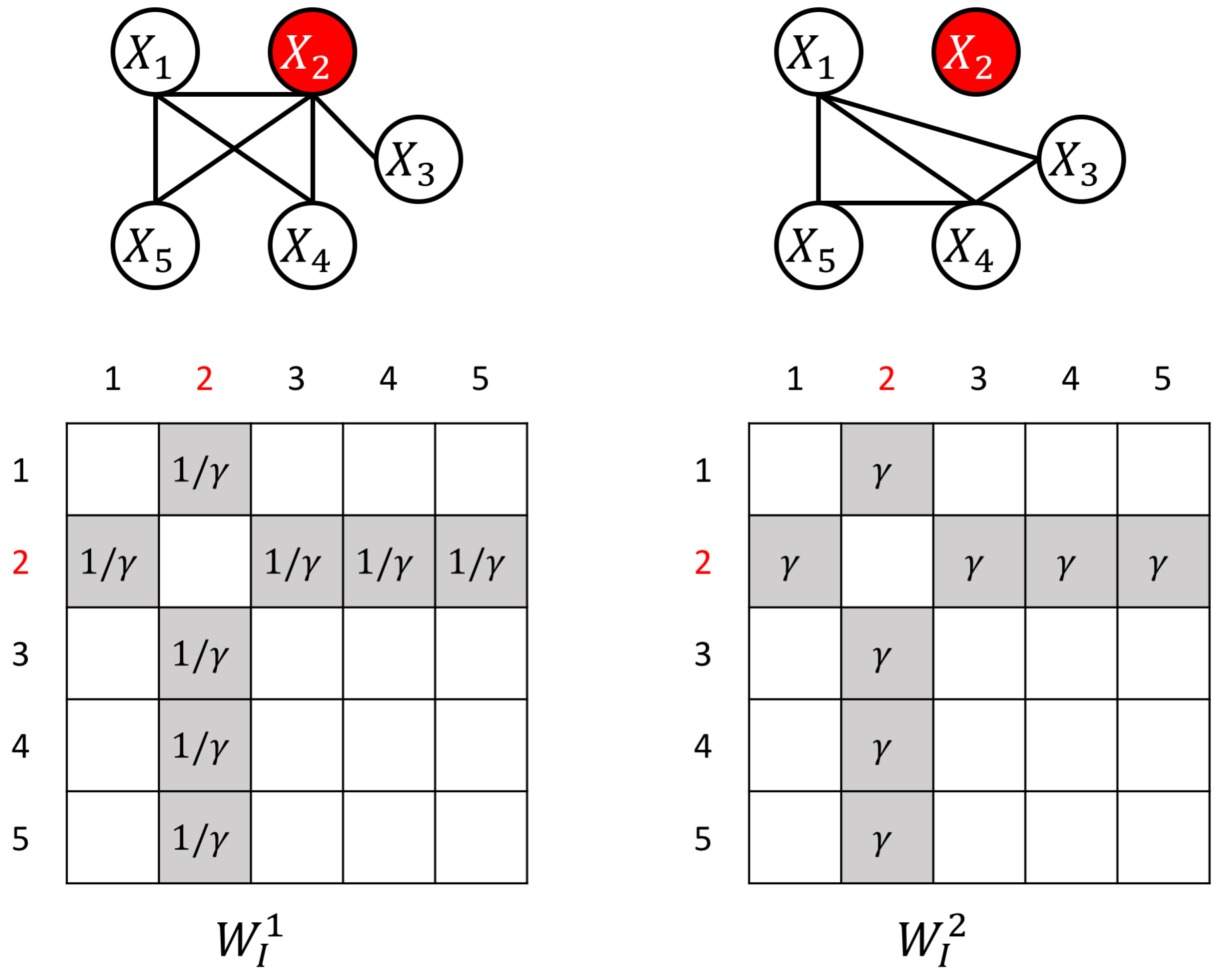
The proposed JEEK estimator provides the flexibility of using ($K+1$) different weight matrices representing the extra knowledge. We try to showcase a few possible designs of the weight matrices, including (but not limited to):
- Spatial or anatomy knowledge about brain regions;
- Knowledge of known co-hub nodes or perturbed nodes;
- Known group information about nodes, such as genes belonging to the same biological pathway or cellular location;
- Using existing known edges as the knowledge, like the known protein interaction databases for discovering gene networks (a semi-supervised setting for such estimations).
We sincerely believe the scalability and flexibility provided by JEEK can make structure learning of joint sGGM feasible in many real-world tasks.
an example W for how to add known group sparity
an example W for how to add known group interactions
an example W for how to add known hub node
an example W for how to add known perturbed-hub node
Citations
@conference{wang2018jeek,
Author = {Wang, Beilun and Sekhon, Arshdeep and Qi, Yanjun},
Booktitle = {Proceedings of The 35th International Conference on Machine Learning (ICML)},
Title = {A Fast and Scalable Joint Estimator for Integrating Additional Knowledge in Learning Multiple Related Sparse Gaussian Graphical Models},
Year = {2018}}
}
Having trouble with our tools? Please contact Beilun and we’ll help you sort it out.
08 Sep 2017
We are updating the R package: simule with one more function: W-SIMULE
install.packages("simule")
library(simule)
demo(wsimule)
Paper: @Arxiv @ NIPS 2017 workshop for Advances in Modeling and Learning Interactions from Complex Data.
Abstract
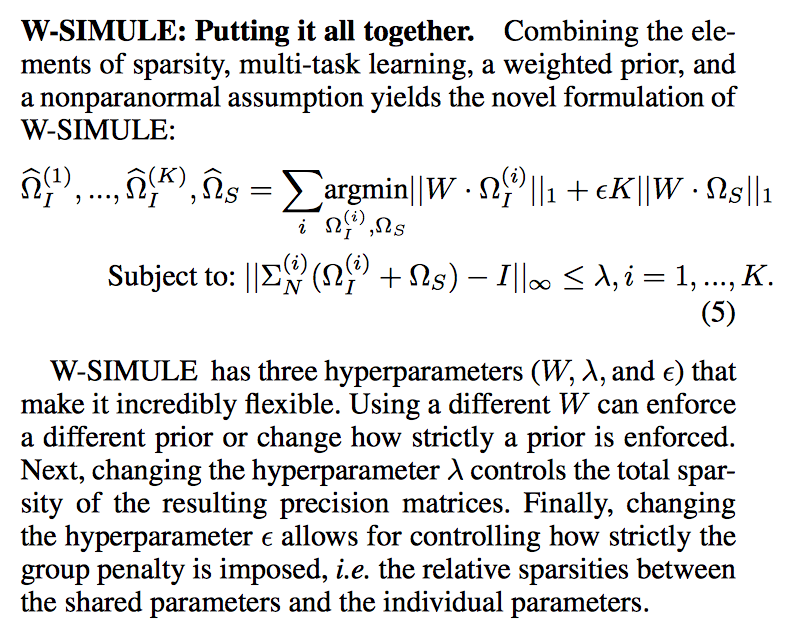
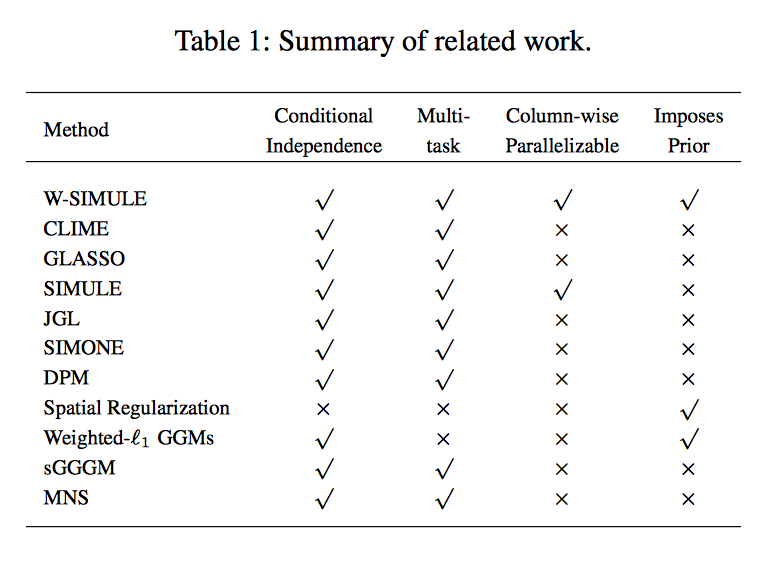
Determining functional brain connectivity is crucial to understanding the brain and neural differences underlying disorders such as autism. Recent studies have used Gaussian graphical models to learn brain connectivity via statistical dependencies across brain regions from neuroimaging. However, previous studies often fail to properly incorporate priors tailored to neuroscience, such as preferring shorter connections. To remedy this problem, the paper here introduces a novel, weighted-ℓ1, multi-task graphical model (W-SIMULE). This model elegantly incorporates a flexible prior, along with a parallelizable formulation. Additionally, W-SIMULE extends the often-used Gaussian assumption, leading to considerable performance increases. Here, applications to fMRI data show that W-SIMULE succeeds in determining functional connectivity in terms of (1) log-likelihood, (2) finding edges that differentiate groups, and (3) classifying different groups based on their connectivity, achieving 58.6\% accuracy on the ABIDE dataset. Having established W-SIMULE’s effectiveness, it links four key areas to autism, all of which are consistent with the literature. Due to its elegant domain adaptivity, W-SIMULE can be readily applied to various data types to effectively estimate connectivity.
Citations
@article{singh2017constrained,
title={A Constrained, Weighted-L1 Minimization Approach for Joint Discovery of Heterogeneous Neural Connectivity Graphs},
author={Singh, Chandan and Wang, Beilun and Qi, Yanjun},
journal={arXiv preprint arXiv:1709.04090},
year={2017}
}
Having trouble with our tools? Please contact Beilun and we’ll help you sort it out.





