FastSK- Fast Sequence Analysis with Gapped String Kernels
01 Jun 2020Title: FastSK: Fast Sequence Analysis with Gapped String Kernels
Paper Bioinformatics
GitHub: https://github.com/QData/FastSK
Talk Slides
Talk video

Abstract
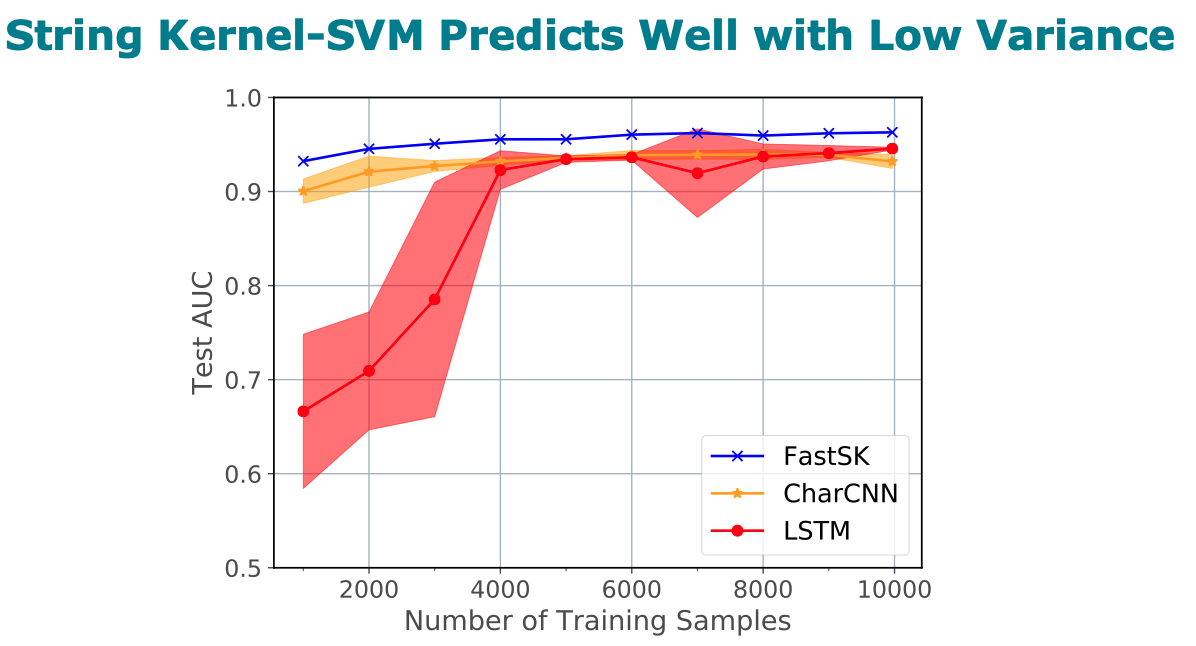
Gapped k-mer kernels with Support Vector Machines (gkm-SVMs) have achieved strong predictive performance on regulatory DNA sequences on modestly-sized training sets. However, existing gkm-SVM algorithms suffer from the slow kernel computation time, as they depend exponentially on the sub-sequence feature-length, number of mismatch positions, and the task’s alphabet size. In this work, we introduce a fast and scalable algorithm for calculating gapped k-mer string kernels. Our method, named FastSK, uses a simplified kernel formulation that decomposes the kernel calculation into a set of independent counting operations over the possible mismatch positions. This simplified decomposition allows us to devise a fast Monte Carlo approximation that rapidly converges. FastSK can scale to much greater feature lengths, allows us to consider more mismatches, and is performant on a variety of sequence analysis tasks. On 10 DNA transcription factor binding site (TFBS) prediction datasets, FastSK consistently matches or outperforms the state-of-the-art gkmSVM-2.0 algorithms in AUC, while achieving average speedups in kernel computation of 100 times and speedups of 800 times for large feature lengths. We further show that FastSK outperforms character-level recurrent and convolutional neural networks across all 10 TFBS tasks. We then extend FastSK to 7 English medical named entity recognition datasets and 10 protein remote homology detection datasets. FastSK consistently matches or outperforms these baselines. Our algorithm is available as a Python package and as C++ source code. (Available for download at https://github.com/Qdata/FastSK/. Install with the command make or pip install)
Citations
@article{10.1093/bioinformatics/btaa817,
author = {Blakely, Derrick and Collins, Eamon and Singh, Ritambhara and Norton, Andrew and Lanchantin, Jack and Qi, Yanjun},
title = "{FastSK: fast sequence analysis with gapped string kernels}",
journal = {Bioinformatics},
volume = {36},
number = {Supplement_2},
pages = {i857-i865},
year = {2020},
month = {12},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btaa817},
url = {https://doi.org/10.1093/bioinformatics/btaa817},
eprint = {https://academic.oup.com/bioinformatics/article-pdf/36/Supplement\_2/i857/35337038/btaa817.pdf},
}
Support or Contact
Having trouble with our tools? Please contact Yanjun Qi and we’ll help you sort it out.