TSK- Transfer String Kernel for Cross-Context DNA-Protein Binding Prediction
15 Jun 2016Tool TSK: Transfer String Kernel for Cross-Context DNA-Protein Binding Prediction
Paper
GitHub
Abstract
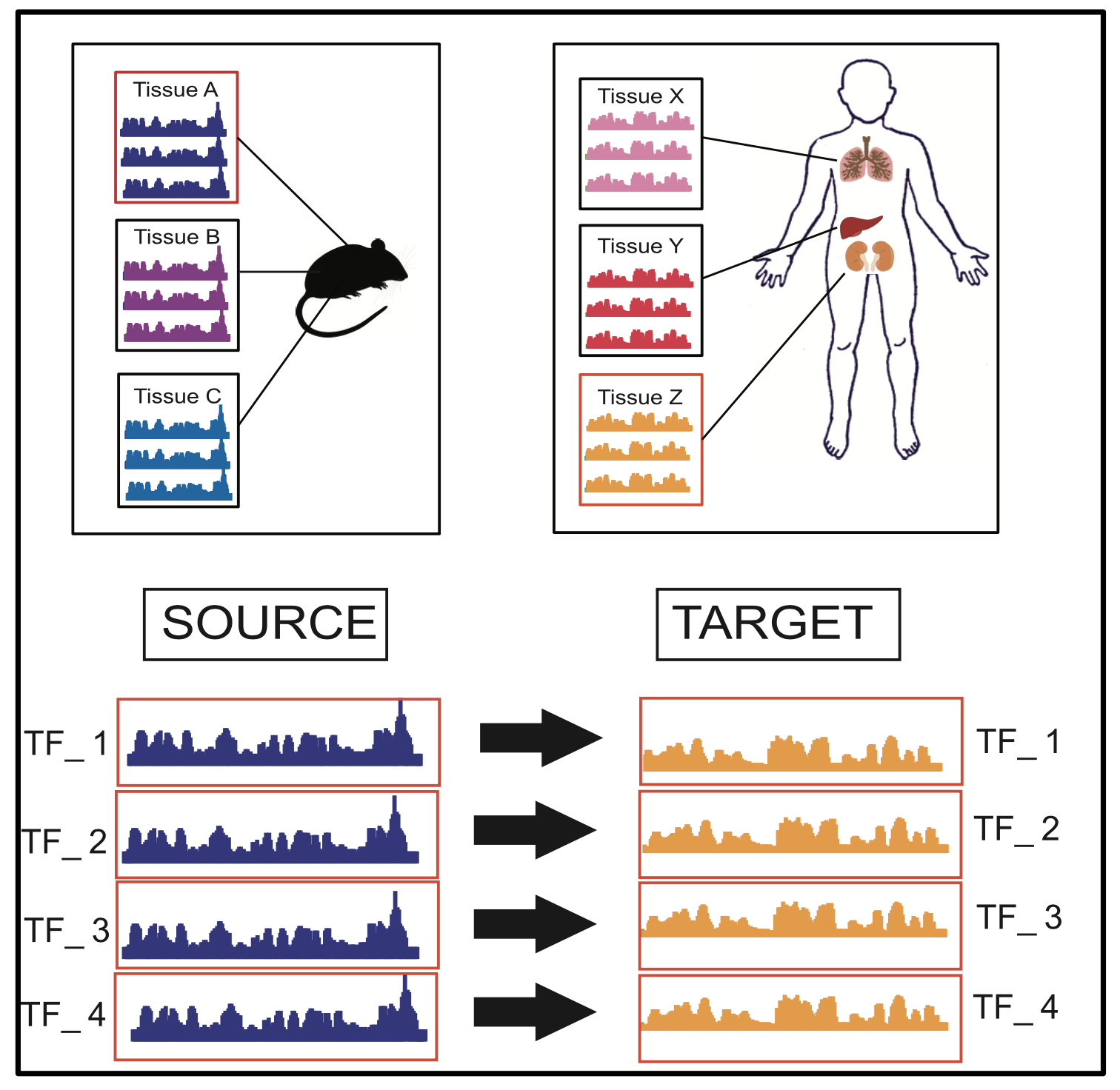
Through sequence-based classification, this paper tries to accurately predict the DNA binding sites of transcription factors (TFs) in an unannotated cellular context. Related methods in the literature fail to perform such predictions accurately, since they do not consider sample distribution shift of sequence segments from an annotated (source) context to an unannotated (target) context. We, therefore, propose a method called “Transfer String Kernel” (TSK) that achieves improved prediction of transcription factor binding site (TFBS) using knowledge transfer via cross-context sample adaptation. TSK maps sequence segments to a high-dimensional feature space using a discriminative mismatch string kernel framework. In this high-dimensional space, labeled examples of the source context are re-weighted so that the revised sample distribution matches the target context more closely. We have experimentally verified TSK for TFBS identifications on fourteen different TFs under a cross-organism setting. We find that TSK consistently outperforms the state-of the-art TFBS tools, especially when working with TFs whose binding sequences are not conserved across contexts. We also demonstrate the generalizability of TSK by showing its cutting-edge performance on a different set of cross-context tasks for the MHC peptide binding predictions.
Citations
@article{singh2016transfer,
title={Transfer String Kernel for Cross-Context DNA-Protein Binding Prediction},
author={Singh, Ritambhara and Lanchantin, Jack and Robins, Gabriel and Qi, Yanjun},
journal={IEEE/ACM Transactions on Computational Biology and Bioinformatics},
year={2016},
publisher={IEEE}
}
Support or Contact
Having trouble with our tools? Please contact Rita and we’ll help you sort it out.